|
|||||||||
|
|
This website was produced as an assignment for Genetics 677 at UW-Madison Spring 2009 Gene Analysis
DNA Sequence DNA Phylogeny Assessment
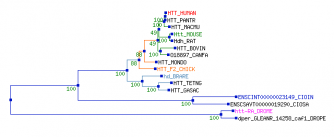
The following is a phylogenetic tree using the HTT gene sequence based on results from Treefam. The HTT human gene appears most closely conserved with Pan troglodytes, abbreviated "HTT_PANTR." This result is expected, and is closely aligned with the protein sequence phylogeny assessment. The most divergent DNA homologues, appearing on the bottom of the graph as "DROME" and "DROPE" represent two species of Drosophila. Interestingly enough, two species of tunicates, Ciona savignyi and Ciona intestinalis (CIOSA and CIOIN, respectively) appeared as the next most similar branch. The branch containing BRARE, TETNG, and GASAC represent three species of bony fishes, including Danio reiro, which also appeared in my protein phylogeny assessment. The rest of the results are more similar and, in my opinion, have a much higher likelihood of sharing common features with the human HTT gene. These include mouse, rat, dog, cow, chicken, and possum HTT genes. This indicates the HTT gene is relatively well conserved, especially amongst mammels. I would therefore expect to see similar protein products in these organisms, along with similar functions in the cell. The Treefam program was quite easy to use, and provided a good deal of graphical information demonstrating the level of similarity in the gene between organisms. PBIL
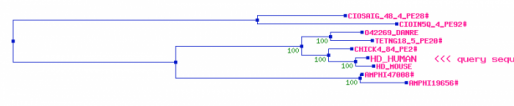
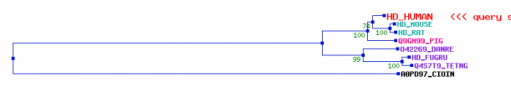
A second website,PBIL, presented a tree using theHOGENOM database which demonstrated a similar phylogenetic arrangement for human huntingtin. The second database used on the site, HOVERGEN, provided a similar arrangment with slightly different similarity scores. The two databases help support the information I gained from Treefam (1,2). HOGENOMHOVERGENCreated by Eric Nickels [email protected] 3/26/2009 Genetics 677 Webpage References: |
||||||||
|
|
|||||||||