|
|||||||||
|
|
This website was produced as an assignment for Genetics 677 at UW-Madison Spring 2009 Gene Ontology
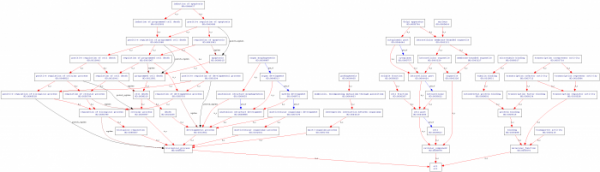
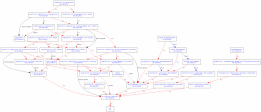
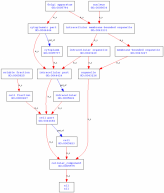
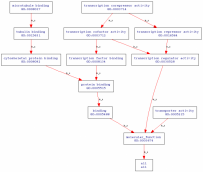
I used two different databases to search for gene ontology terms for human HTT. Other databases I searched did not have a human HTT gene annotated. AmiGOThe AmiGO database listed 11 ontology terms under human HTT, four each for biological processes and molecular function and three for cellular component. The site provides a simple method to create trees showing linkage between the different terms. The results for each compenent are shown below. Additionally, I created a tree showing linkage between all 11 terms, shown directly below. Biological process
The biological process terms indicate the role the gene plays in the larger biological framework of the cell. Of the terms listed on AmiGO, apoptosis and pathogenesis at first appeared the most significant in describing the role of HTT in Huntington's disease. It has been demonstrated that the mutant form of huntingtin eventually leads to apoptosis in neurons (1). The role in organ morphogenesis was seen mice knockouts for huntingtin (2). GO Terms Cellular component
The cellular component terms describe the locations in the cell in which the gene is actively functioning. I was not surprised to see both cytoplasm and the nucleus listed, as huntingtin is described as active in both in primary papers I previously came across. References for both are listed on AmiGO. The soluble fraction term essentially indicates the protein is found in the water soluble components of the cell, which includes the cytoplasm and nucleus. The golgi apparatus listing led me to a research paper describing the interaction of huntingtin with optineurin, which maintains the shape of the golgi apparatus (3). GO Terms Molecular function
Molecular function terms describe the precise role the protein plays in the cell on the molecular level. The GO terms for molecular function included three similar terms in microtuble binding, protein binding, and transporter activity, all of which point to huntingtin actively taking part in transport in the cell. The reference listed under transporter activity describe the role of huntingin as anchoring microtubules or transporting mitochondria, vesicles, or other organelles (4). The transcriptional corepressor reference describes an interaction of mutant huntingtin with p53, repressing p53-driven promoters p21 and MDR-1 (5). Furthermore, mutant huntingtin binds CREB binding protein, which overall results in reduced control of transcription (5). GO Terms GOALL
The GOALL database listed 55 GO terms under the gene name HTT for humans. Of the biological processes, some corresponded with functions I would consider highly connected to the nature of Huntington's disease, including brain development and locomotory behavior. Others were surprising, such as iron ion homeostasis. The cellular component and molecular function terms followed the same pattern, with more terms listed for each compared to AmiGo. Subcellular Localization
Based upon my results for gene ontology, I used a sequence search in the pTARGET database to determine subcellular locations of the protein based upon its structure. pTARGET demonstrated with relatively high confidence that huntingtin locates to the cytoplasm based upon its structure, with a confidence value of 87.6%. I found this interesting, as the protein sequence I used was the full huntngtin (3,144 aa). The localization to the nucleus listed in my GO terms is proposed to occur only when the protein is cleaved. AnalysisThe AmiGO program provided what I considered the most useful information regarding the gene ontology of huntingtin. I thought the tree creation aspect of the database was a useful way to manipulate the ontology terms to visualize how the protein's function is mapped out. Also, the direct link to the primary papers accounting for the papers provided useful information and a good way to build up background knowledge on each specific aspect of the protein. Overall, the AmiGO program outlined the importance of huntingtin in transport activities, with the possiblity of function in both the cytoplasm and the nucleus. This database would be useful to gather intial information about a particular gene or protein to gain an understanding of its function. GO ALL, in contrast, provided more terms, though the large number made the characterization of the actual function of the protein more difficult. GO ALL would probably be more useful to search for more unique functions of a protein. References: Created by Eric Nickels [email protected] 5/9/2009 Genetics 677 Webpage |
||||||||
|
|
|||||||||