|
|||||||||
|
|
This page was created as a project for Genetics 677 at UW-Madison in the spring of 2009. Protein Interactions
String database
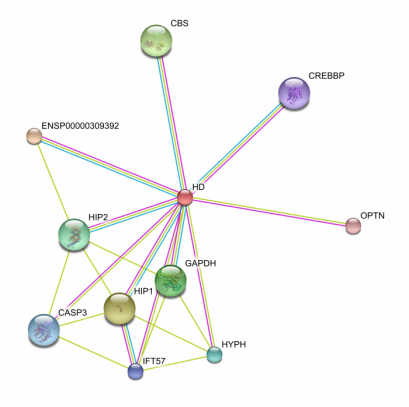
The String database provided an interaction web for the human huntingtin protein, shown below, as well as D. melanogaster and Mus Musculus. A table documenting the interaction score for each protein and the manner its interaction was determined is also demonstrated below. The most highly scoring protiens (at 0.999) HAP1 and HIP1, represent huntingtin associating protein and huntingtin interacting protein, respectively. The function of these two proteins is, clearly, quite determined by the huntingtin protein. A more thorough utilization of STRING
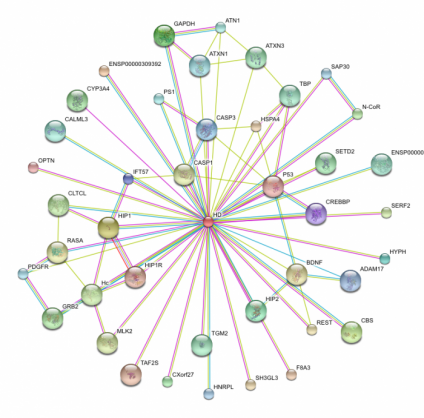
The default settings provide the top ten interactions by confidence level in their itneraction with huntigntin. Thus, I expanded my search to see how many interactions huntingtin is implicated in. First searching under the medium confidence level setting (0.400 or greater) I retrieved well over 100 interactions. Narrowing this down to the top confidence level (0.900 or greater) I still retrieved 41 different interactions. These 41 proteins demonstrate some of the key pathways which the mutant huntingtin protein may be involved in. These protein interactions are key to determining how huntingtin functions in the cell. For instance, the STRING diagram demonstrates an interaction with BDNF, or brain derived neurotrophic factor precursor, indicates that the pathway for this trophic factor's utilziation may be blocked in the disease state. Some studies indicate that restoring BDNF in mouse Huntington's disease models may be an effective treatment for preventing neurodegradation (2). P53, as described below in the KEGG pathway, maintains and obvious importance in cell survival and the induction of apoptosis, which may demonstrate an insight into how huntingtin actually induces cell death. A calmodulin protein appeared in this web, which I found interesting due to the fact that the KEGG diagram below indicates that calcium ion homeostasis is disrupted in the Huntington's disease state, although this particular protein, CaM, listed as ENSP00000349467, is not part of this pathway. Mutant huntingtin is actually capable of binding calmodulin (3). Blocking the ability of hunitngitn to calmodulin has been proposed as a method ot protect neurons from death (3). This overwhelming number of interactions perhaps best indicates how expansive the action of the mutant form of huntingtin is, as well as the complexity and difficulty of determining the basis of pathogenesis in Huntington's disease. Other Species
The Mus musculus homolog, hdh or huntingtin disease homolog, showed a much greater similarity in its interactions to human huntingtin, which is expected due to the high level of similarity between the proteins and the relatively relatedness of the species. Hdh interacts with HAP1, HIP1 and HIP2, as well as other interactions equivalent to the human form. The Pan troglodytes homolog was somewhat less intricate in its interaction web, altough a high level of similarity is seen between this species and humans in the proteins huntingtin interacts with. It appears the interactions of proteins linked to huntingtin, and not the huntingtin interactions themselves, are either less full annotated or do not interact in P. troglodytes, although the former explanation seems more likely. Osprey Program
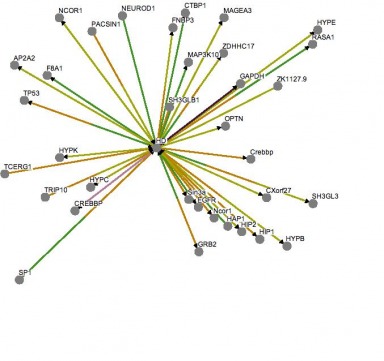
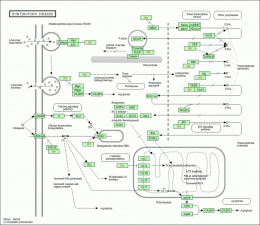
The Osprey program allows the creation of interaction maps from uploaded data. I found a dataset on Biogrid corresponding to human huntingtin interactions. Below is the interaction web created by the Osprey program on this data. The interaction web lists a greater number of interactions for HD (center) than the STRING program. Overall, it appears the core constituents of the web, similar to those found in STRING, relate the general function of mutant huntingtin to a vesicle transport protein and some type of interaction in the nucleus with DNA binding proteins, such as CREBBP. Above: HD interaction web generated by Osprey using Biogrid data. KEGGKEGG provides detailed pathway diagrams of protein interactions. The database listed pathway diagrams for huntingtin for a variety of species, including other species I previously described as having homologous proteins, though the diagrams appear identical for all of the listed species. The human version is extensive in detail, involving numerous pathways, which perhaps indicates the lack of conclusive evidence for the specific role of huntingtin in neurons. Diagrammed within a striatal medium spiny neuron, the main processes involved include abnormal clathrin mediated endocytosis, altered vescile transport, transcriptional repression, abnormal calcium homostasis, mitochondrial dysfunction, increased caspase activity and increased apoptosis. The diagram shows interactions with many of the proteins described in interaction webs above, including HIP1 and HAP1. This diagram in particular is extensively detailed, and gives a better indication of the actual cellular processes altered in the interactions of mutant huntingtin. One advantage to using KEGG is the ability to select individual proteins in the pathway to link to databases listinngs. Above: KEGG diagram of mutant huntingtin interactions Panther
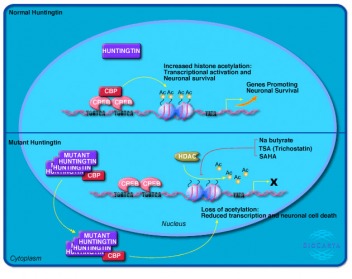
The Panther program provided another pathway diagram for human huntingtin. The Panther diagram was very similar to KEGG, although Panther has a more interactive interface. The diagram is essentially the same as the KEGG pathway, althought it appears KEGG contained a slightly larger amount of interactions. BiocartaThe Biocarta site listed a pathway for the wild type and mutant huntingtin protein in the nucleus. The normal huntingtin function is not extensively diagrammed, rather Biocarta demonstrates the wild type protein does not interfere with CBP binding to CREB. CBP, or CREB binding protein, increases basal transcription levels and acetylates histones of neuronal genes, which leads to higher levels of transcription overall. The mutant huntingtin, which forms aggregates both within the nucleus and cytoplasm, binds CBP. This results in an overal decrease in histone acetylation, which decreases transcription. Above: Biocarta huntingtin protein pathway for normal and mutant protein. Overall Analysis
The variety of databases listing protein interaction diagrams and webs clearly demonstrates the current state of understanding of the huntingtin protein's function in the cell, which is to say there is a massive amount of proposed data while the actual mechanisms involved in the pathogenesis of the disease may still be misunderstood. While the mechanism may not be entirely known, the protein interaction diagrams provide great insight into the possible effects of the mutant huntingtin protein on the cell. The Biocarta pathway, while relatively simple in its diagram, draws a parallel to a microarray experiment I analyzed in which overall transcription was reduced in striatal neuronal cells. The two larger pathway diagrams, KEGG and Panther, describe the implications of the mutant huntingtin protein in a more widespread cellular manner; we see the effect of the protein aggregates is much more complex than the one-step mechanism portrayed in Biocarta. The STRING database supports the two larger pathway diagrams, as the total number of protein interactions is extensive. In the other species with documented interaction webs on STRING, it appears the interactions are generally similar, which indicates a specific role for huntingtin that is equivalent at least throughout the mammalian lineage. The databases available for protein interactions are very useful in regards to understanding the known actions of huntingtin, as it is well annotated in most databases, and is a very quick way to assess the overall action of the protein in the cell. References: Created by Eric Nickels [email protected] 5/8/2009 Genetics 677 Webpage |
||||||||
|
|
|||||||||