|
|||||||||
|
|
This website was created as a project for Genetics 677 at UW-Madison in the Spring of 2009 Small Molecule Interactions
I searched several databases to determine if any significant data exists indicating the interaction between the huntingtin protein and small molecules. The first database I used was PubChem. Small molecules shown to aid in the clearance of mutant huntingtin aggregates
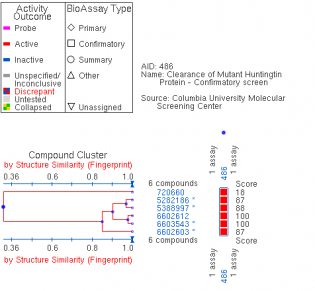
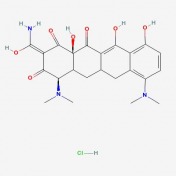
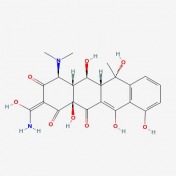
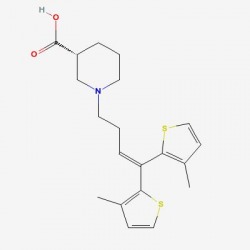
One of the main molecular markers of mutant huntingtin is the aggregation of cleaved protein molecules. These aggregates contribute to the pathological symptoms of the disease. A study by Columbia University , denoted AID 853, tested the ability of small molecules to bind and aid in the removal of aggregated hungtingtin. This was a high throughput, primary screen. Of the 46,735 molecules against mutant huntingtin, 195 were found to have strong interactions. A complete tree of these molecules can be seen here, as related to biological activity of the molecules. The researchers then conducted a confirmatory screen, AID 486, using 92 of these molecules. Structure/Activity analysis of the six active compounds identified by the confirmatory screen AID 486. The six structures are described below. Chlorotetracycline (SID 855713) MLS000028422 (SID 855598) Tetracycline (SID 855950) Minocycline (SID 855650) Oxytetracycline (SID 855768) MLS000061862 (SID 4264628) ResultsPubChem provided a simple, straightforward method of searching for chemical screens involving huntingtin. I found the process of narrowing the search field in the high throughput analysis, throughout a total of three screens, interesting in that the procedural modifications in each screen turned up different activity results for each molecule. The tetracycline derivatives, the highest scoring compounds in the confirmatory screen, were not the highest scoring molecules on the initial screen. The use of tetracycline derivatives in the treatment of Huntington's disease has been examined further in experiments using the R6/2 mutant huntingtin expressing mouse model. One study conducted by Mievis et al. demonstrated minocycline is ineffiecienty in treating Huntington's disease in vivo in the R6/2 mouse (1). Overall, it does not appear there are any studies conducted in living models of Huntington's disease in which tetracycline derivatives how any therapeutic effect. However, studies using other drugs not included in the above screen show therapeutic effects in mouse models. Masuda et al. demonstrated the neuroprotective effects of tiagabine in Huntingon's disease mouse models, including R6/2 (2). This prompted a search on PubChem for tiagabine, the results of which follow. Tiagabine, above, was found to limit neuronal cell death in hunting mutant cells by Masuda et al. The neuroprotective effect of tiagabine
I searched PubMed for tiagabine and found information regarding the physiological action of the drug. Tiagabine is implicated in three different pharmacological actions, listed as an anticonvulsant, a GABA neurotransmitter rececptor agonist, and a neurotransmitter uptake inhibitor. The latter two actions indicate the drug prolongs action of neurotransmitter release from neurons. By blocking neurotransmitter uptake into the axon terminal of the presynaptic neuron, tiagabine prolongs the stimulatory action of the neurotransmitter once released into th synaptic cleft. As a GABA agonist, tiagabine activates post-synaptic GABA receptors. The anticonvulsant action of tiagabine may also be significant in Huntington's disease in the treatment of the characteristic chorea experienced by patients affected by the disease. Tiagabine was indentifed in the high throughput screen AID 1471, attributed to NCGC. The research was conducted on a cell based model of Huntington's disease, in which a GFP protein was attached to the mutant form of exon 1 of huntingtin. When aggregates of huntingtin formed, the GFP signal became more detectable. Interacting drugs were able to reduce the amount of GFP in the cell and resulted in greater cell survival. Other databases
Other databases I searched, including ChemBank and DrugBank, did not return any results or screens for the huntingtin protein. References Created by Eric Nickels [email protected] 4/17/2009 Genetics 677 Webpage |
||||||||
|
|
|||||||||