|
|||||||||
|
|
This site was created as a project for Genetics 677 at UW-Madison in spring 2009 Domain homology
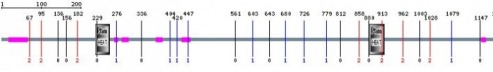
In order to determine the functional similarity of the species containing homologs of the human huntingtin protein, I conducted a SMART analysis of the sequences to search for similar domains. Below are the results for each species. I used the first 2,000 amino acids for this search, as it was the size limit for SMART, although an analysis of the final 2,000 amino acids did not return significant results for my project. I also used images showing only around the first 1,000 amino acids, although amino acids 1,000 to 2,000 did not return any significant results. Homo sapiens Pan troglodytes Mus musculus Rattus norvegicus Bos tarus Gallus gallus Danio rerio Looking at the similarity between all of the homologs and HEAT repeat domains, two key aspects of the protein are appearent. First, the polyglutamine region, demonstrated by the pink region to the N-terminus (near the left side) of the protein, is different in each species, with the greatest length seen in humans and chimps. The zebrafish does not seem to have any polyglutamine region assessed by SMART. Second, the HEAT repeat domains, although indicated to be conserved amoungst these species, differs within each. We see some organisms contain 2 heat repeat domains, while some, including humans, contain only one. This may be accounted for simply by the relative unique structure of the protein and perhaps an inadequate definition of the sequence of HEAT repeats. If, for instance, the HEAT domains differ by a few amino acids, it perhaps would not appear on these diagrams. Even so, it is likely that these proteins, based upon these domains, should share a very similar function in each organism. The length of the polyQ region indicates the pathological effects of the protein are probably not similar in these organisms. |
||||||||
|
|
|||||||||