|
||||||||||
|
|
This website was produced as an assignment for Genetics 677 at UW-Madison Spring 2009 Protein Sequence and Homology
Human Sequence of Huntingtin
The protein sequence used in this project was obtained from the NCBI site (NP 002102) The huntingtin protein is 3,144 amino acids in length. From a search on ExPASy, I found the isoelectric point of huntingtin is 5.81 and the molecular weight 347,859.56 Da, based upon this sequence. HomologsIn order to determine the closest protein homologs to human huntingtin, I first conducted a general search through Homologene to develop a list of other organisms potentially expressing a similar protein. The following list contains these proteins found in other species homologous to the human huntingtin protein. The proteins are listed by their corresponding gene name. Click on the protein's accession number to view amino acid sequence data in FASTA format from Entrez Protein. After generating a potential list, I attempted to compare the level of similarity between human huntingtin and the homologs through Blastp,a protein-protein algorithm. Pan troglodytes(chimpanzee)
solute carrier family 6 protein Canis lupus familiaris (dog)
solute carrier family 6 protein Bos taurus (cattle)
Huntingtin Danio rerio (zebrafish)
Huntingtin Mus musculus (mouse)
Huntingtin Rattus norvegicus (rat)
Huntingtin Gallus gallus (chicken)
Huntingtin Sequence Alignments
I used two programs to assess the similarity between all of the homologous sequences for huntingtin. The results below show a relatively high level of conservation among the homologues. The t-coffee alignment gives a colored key to similarity scores for each amino acid. I also ran a muscle alignment, given below. The alignments show a relatively high level of conservation in the amino acid sequences of the homologs.
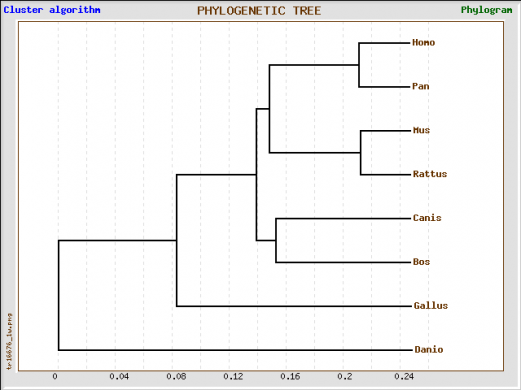
Phylogenetic Tree Analysis
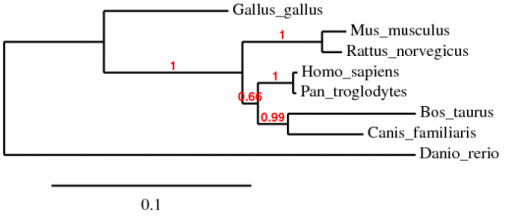
I constructed the above phylogenetic tree using the Treetop program, analyzing for homology in the huntingtin protein homoogues for the species listed above. Using the standard settings on the program, I generated a tree showing the greatest similarity of the human form of huntingtin to chimpanzees. The greatest divergence is between the human and the zebrafish forms. I then performed a second phylogenetic analysis using Phylogeny.fr on "One-Click" settings. The results were similar to those of Treetop. We see the human form of hungtingtin is very similar to the chimpanzee form. Scores indicate relative similarities for specific branches-thus, with a score of 1, the Homo sapiens and Pan troglodyes huntingtin homologues are quite similar, almost identical. Overall Analysis
The BLAST protein sequence scores show there is a high level of similarity between the homologs in the different organisms. The scores on the BLAST program match up well with my phylogenetic analysis of the homologs. I found the BLAST program to be informative and simple to use, although the depth of information in the results is not in particular that great. The different sequence alignment programs further supported these results. I found the information from T-COFFEE to be the most useful, in the sense that all 4 programs I used brought back the same results, while T-COFFEE was the only program that represented the data graphically (in color). The motif search programs were difficult to use with huntingtin - the expected HEAT repeats were not immediately found by any of the programs. The relative scores for the HEAT repeats in the programs matched those of low-complexity regions and other undefined motifs. Thus, I wasn't able to put a lot of confidence in my results from the motif programs, although the literature on huntingtin confirmed the presence of HEAT repeats. I believe developing greater familiarity with these programs will allow me to gain better information in the future and will be more informative in my analysis of huntingtin. Created by Eric Nickels [email protected] 5/8/2009 Genetics 677 Webpage |
|||||||||
|
|
||||||||||